File:Uusia koronavirustapauksia suomessa fourier ennuste 1.svg

Original file (SVG file, nominally 892 × 520 pixels, file size: 41 KB)
Captions
Captions
Summary
[edit]| DescriptionUusia koronavirustapauksia suomessa fourier ennuste 1.svg |
Suomi: Uusia koronavirustapauksia suomessa - Fourier ennuste |
| Date | |
| Source | Own work |
| Author | Merikanto |
Python3 code
- COVID-19 Finland cases short-term forecast
- Python 3 script
- Input from internet site: cases.
- version 0000.0006
- 15.4.2022
-
import locale
import math as math
import numpy as np
import pandas as pd
import statistics
import matplotlib.pyplot as plt
from numpy import fft
from scipy import interpolate
import scipy.signal
from scipy.signal import savgol_filter
from scipy.interpolate import UnivariateSpline
from scipy.interpolate import pchip_interpolate
from scipy import stats
from scipy.optimize import curve_fit
from datetime import datetime, timedelta
from datetime import date
import matplotlib.dates as mdates
import matplotlib.ticker as ticker
from matplotlib.ticker import ScalarFormatter
from matplotlib.ticker import (MultipleLocator, FormatStrFormatter,
AutoMinorLocator, MaxNLocator)
- from mizani.breaks import date_breaks, minor_breaks
- from mizani.formatters import date_format
from bs4 import BeautifulSoup
import requests
- asetukset parameters
- pohjadatan rajat: limits of base data
today = date.today()
- print("Today's date:", today)
daybeforeyesterday = datetime.today() - timedelta(days=2)
paiva1="2022-03-01"
paiva2="2022-04-05"
- paiva2=daybeforeyesterday
- ennusteen rajat: forecast limits
taika1=paiva2
taika2="2022-07-01"
- y-akselin rajat
ymax1=12000
ymax2=50
vormat1='%Y-%m-%d'
- locale.setlocale(locale.LC_TIME, "fi_FI")
def func(x, a, b):
return (a*x+b)
def fourierExtrapolation(x, n_predict):
n = x.size
n_harm = 10 # number of harmonics in model
t = np.arange(0, n)
p = np.polyfit(t, x, 1) # find linear trend in x
x_notrend = x - p[0] * t # detrended x
x_freqdom = fft.fft(x_notrend) # detrended x in frequency domain
f = fft.fftfreq(n) # frequencies
indexes = range(n)
# sort indexes by frequency, lower -> higher
#indexes.sort(key = lambda i: np.absolute(f[i]))
t = np.arange(0, n + n_predict)
restored_sig = np.zeros(t.size)
for i in indexes[:1 + n_harm * 2]:
ampli = np.absolute(x_freqdom[i]) / n # amplitude
phase = np.angle(x_freqdom[i]) # phase
restored_sig += ampli * np.cos(2 * np.pi * f[i] * t + phase)
return restored_sig + p[0] * t
def cut_by_dates(dfx, start_date, end_date):
mask = (dfx['Date'] > start_date) & (dfx['Date'] <= end_date)
dfx2 = dfx.loc[mask]
#print(dfx2)
return(dfx2)
def load_country_cases(maa):
dfin = pd.read_csv('https://datahub.io/core/covid-19/r/countries-aggregated.csv', parse_dates=['Date'])
countries = [maa]
dfin = dfin[dfin['Country'].isin(countries)]
#print (head(dfin))
#quit(-1)
selected_columns = dfin"Date", "Confirmed", "Recovered", "Deaths"
df2 = selected_columns.copy()
df=df2
len1=len(df["Date"])
aktiv2= [None] * len1
for n in range(0,len1-1):
aktiv2[n]=0
dates=df['Date']
rekov1=df['Recovered']
konf1=df['Confirmed']
death1=df['Deaths']
#print(dates)
spanni=6
#print(rekov1)
#quit(-1)
rulla = rekov1.rolling(window=spanni).mean()
rulla2 = rulla.rolling(window=spanni).mean()
tulosrulla=rulla2
tulosrulla= tulosrulla.replace(np.nan, 0)
tulosrulla=np.array(tulosrulla).astype(int)
rulla2=tulosrulla
x=np.linspace(0,len1,len1);
#print("kupla")
#print(tulosrulla)
#print(konf1)
#print(death1)
#print(aktiv2)
konf1=np.array(konf1).astype(int)
death1=np.array(death1).astype(int)
#print(konf1)
#quit(-1)
for n in range(0,(len1-1)):
#print("luzmu")
rulla2[n]=tulosrulla[n]
#print ("luzmu2")
#aktiv2[n]=konf1[n]-death1[n]-rulla2[n]
aktiv2[n]=konf1[n]
#print(rulla2[n])
#quit(-1)
#aktiv3=np.array(aktiv2).astype(int)
dailycases1= [0] * len1
dailydeaths1= [0] * len1
for n in range(1,(len1-1)):
dailycases1[n]=konf1[n]-konf1[n-1]
if (dailycases1[n]<0): dailycases1[n]=0
for n in range(1,(len1-1)):
dailydeaths1[n]=death1[n]-death1[n-1]
if (dailydeaths1[n]<0): dailydeaths1[n]=0
#quit(-1)
df.insert (2, "Daily_Cases", dailycases1)
df.insert (3, "Daily_Deaths", dailydeaths1)
df['ActiveEst']=aktiv2
#print (df)
dfout = df'Date', 'Confirmed','Deaths','Recovered', 'ActiveEst','Daily_Cases','Daily_Deaths'
#print(df)
#print(dfout)
print(".")
return(dfout)
def load_fin_wiki_data():
url="https://fi.wikipedia.org/wiki/Suomen_koronaviruspandemian_aikajana"
response = requests.get(url)
soup = BeautifulSoup(response.text, 'lxml')
table = soup.find_all('table')[0] # Grab the first table
df = pd.read_html(str(table))[0]
#print(df)
#Päivä Tapauksia Uusia tapauksia Sairaalassa Teholla Kuolleita Uusia kuolleita Toipuneita
df2 = df'Tapauksia','Uusia tapauksia','Sairaalassa','Teholla','Kuolleita','Uusia kuolleita','Toipuneita'
kaikkiatapauksia=df['Tapauksia']
toipuneita=df['Toipuneita']
uusiatapauksia=df['Uusia tapauksia']
sairaalassa=df['Sairaalassa']
teholla=df['Teholla']
kuolleita=df['Kuolleita']
uusiakuolleita=df['Uusia kuolleita']
len1=len(kaikkiatapauksia)
kaikkiatapauksia2=[]
toipuneita2=[]
uusiatapauksia2=[]
sairaalassa2=[]
teholla2=[]
kuolleita2=[]
uusiakuolleita2=[]
for n in range(0,len1):
elem0=kaikkiatapauksia[n]
elem1 = .join(c for c in elem0 if c.isdigit())
elem2=int(elem1)
kaikkiatapauksia2.append(elem2)
elem0=toipuneita[n]
elem1 = .join(c for c in elem0 if c.isdigit())
#toipuneita2.append(int(elem1))
if (elem1!=): toipuneita2.append(int(elem1))
else: toipuneita2.append(0)
elem0=uusiatapauksia[n]
elem1 = .join(c for c in elem0 if c.isdigit())
uusiatapauksia2.append(int(elem1))
elem0=sairaalassa[n]
#elem1 = .join(c for c in elem0 if c.isdigit())
sairaalassa2.append(int(elem0))
elem0=teholla[n]
#elem1 = .join(c for c in elem0 if c.isdigit())
teholla2.append(int(elem0))
elem0=kuolleita[n]
#elem1 = .join(c for c in elem0 if c.isdigit())
kuolleita2.append(int(elem0))
elem0=uusiakuolleita[n]
#elem1 = .join(c for c in elem0 if c.isdigit())
uusiakuolleita2.append(int(elem0))
#kaikkiatapauksia3=np.array(kaikkiatapauksia2).astype(int)
#print("---")
#print(kaikkiatapauksia2)
#print(toipuneita2)
kaikkiatapauksia3=np.array(kaikkiatapauksia2).astype(int)
toipuneita3=np.array(toipuneita2).astype(int)
uusiatapauksia3=np.array(uusiatapauksia2).astype(int)
sairaalassa3=np.array(sairaalassa2).astype(int)
teholla3=np.array(teholla2).astype(int)
kuolleita3=np.array(kuolleita2) .astype(int)
uusiakuolleita3=np.array(uusiakuolleita2).astype(int)
for n in range(1,len1):
toipu0=toipuneita3[n]
if (toipu0==0):
paikka=n
toipui=toipu1
break
toipu1=toipu0
## oletus: toipuu suureksi osaksi akuutista vaiheesta 3 viikossa
## todellisuudessa yli 60% kärsii ainakin yhdestä pitkäkestoisesta oireesta
for n in range(paikka,len1):
toipui=toipui+uusiatapauksia3[n-21]-uusiakuolleita3[n]
toipuneita3[n]=toipui
#print(toipuneita3[n])
napapaiva1 = np.datetime64("2020-04-01")
timedelta1= np.timedelta64(len(kaikkiatapauksia3),'D')
napapaiva2 = napapaiva1+timedelta1
#dada1 = np.linspace(napapaiva1.astype('f8'), napapaiva2.astype('f8'), dtype='<M8[D]')
dada1 = pd.date_range(napapaiva1, napapaiva2, periods=len(kaikkiatapauksia3)).to_pydatetime()
#print(dada1)
data = {'Date':dada1,
'Kaikkia tapauksia':kaikkiatapauksia3,
"Uusia tapauksia":uusiatapauksia3,
"Sairaalassa":sairaalassa3,
"Teholla":teholla3,
"Kuolleita":kuolleita3,
"Uusiakuolleita":uusiakuolleita3,
"Toipuneita":toipuneita3
}
df2 = pd.DataFrame(data)
#print(kaikkiatapauksia3)
#print ("Fin wiki data.")
return(df2)
def get_solanpaa_fi_data():
url="https://covid19.solanpaa.fi/data/fin_cases.json"
response = requests.get(url,allow_redirects=True)
open('solanpaa_fi.json', 'w').write(response.text)
with open('solanpaa_fi.json') as f:
sola1=pd.read_json(f)
sola1_top = sola1.head()
#print (sola1_top)
#Rt […]
#Rt_lower […]
#Rt_upper […]
#Rt_lower50 […]
#Rt_upper50 […]
#Rt_lower90 […]
#Rt_upper90 […]
#new_cases_uks […]
#new_cases_uks_lower50 […]
#new_cases_uks_upper50 […]
#new_cases_uks_lower90 […]
#new_cases_uks_upper90 […]
#new_cases_uks_lower […]
#new_cases_uks_upper […]
dada1=sola1["date"]
casa1=sola1["cases"]
death1=sola1["deaths"]
newcasa1=sola1["new_cases"]
newdeath1=sola1["new_deaths"]
hosp1=sola1["hospitalized"]
icu1=sola1["in_icu"]
rt=sola1["Rt"]
newcasauks=sola1["new_cases_uks"]
print(dada1)
len1=len(dada1)
toipuneita=np.zeros(len1)
toipuneita=toipuneita*0
aktiivisia=toipuneita
toipui=0
for n in range(0,len1):
if(n>21):
toipui=toipui+newcasa1[n-21]-newdeath1[n]
toipuneita[n]=toipui
aktiivisia=casa1-toipuneita-death1
#print(toipuneita)
#print("KKKK")
#print (dada1[0+21])
data = {'Date':dada1,
'Tapauksia':casa1,
'Kuolemia':death1,
'Sairaalassa':hosp1,
'Teholla':icu1,
'Uusia_tapauksia':newcasa1,
'Uusia_kuolemia':newdeath1,
'R':rt,
'Uusia_tapauksia_ennuste':newcasauks,
'Toipuneita':toipuneita,
'Aktiivisia':aktiivisia,
}
df = pd.DataFrame(data)
return(df)
def calcu_fourier_1(cases,days3):
hattuja=7
fatticache=[]
for n in range(3, hattuja-1,2):
lugu1=(lencases1-n)*-1
casest=cases[lugu1:]
sulen1=len2-lencases1
casest2=np.pad(casest, ((n), 0), 'constant')
casest3=savgol_filter(cases, n, 2)
casest3=cases
yfat1=fourierExtrapolation(casest3,sulen1)
fatit1=yfat1.astype(int)
fatticache.append(fatit1)
hattuja2=len(fatticache)
for n in range(0, hattuja2):
fatit1=fatticache[n]
fotticache1=[]
for n in range(0,len3-1):
fotti=[]
for m in range(0, hattuja2):
fatit1=fatticache[m]
elem1=fatit1[n]
fotti.append(elem1)
fotti1=np.array(fotti)
fotticache1.append(fotti1)
fottitaulu00=np.array(fotticache1)
fottitaulu=fottitaulu00.reshape(len3-1, hattuja2)
devis00=[]
meas00=[]
for n in range(0,len3-1):
sarake=fottitaulu[n]
devis00.append(statistics.stdev(sarake))
meas00.append(statistics.mean(sarake))
devis=np.array(devis00)
meas=np.array(meas00)
return(meas)
-
- df=load_country_cases('Finland')
- df2=cut_by_dates(df, paiva1,paiva2)
df=get_solanpaa_fi_data()
- df=load_fin_wiki_data()
- print(df)
df2=cut_by_dates(df, paiva1,paiva2)
print(df2)
print("...")
- quit(-1)
- dates0=df2['Date']
- cases0=df2['Daily_Cases']
dates0=df2['Date']
- cases0=df2['Daily_Cases']
dailycases1=df2['Uusia_tapauksia']
dailydeaths1=df2['Uusia_kuolemia']
cases0=dailycases1
- quit(-1)
cases=np.array(cases0).astype(int)
- dates=np.array(dates0).to_pydatetime()
dates=dates0
savgol=savgol_filter(cases, 7, 2)
casesminusavgol=cases-savgol
stdev=statistics.stdev(casesminusavgol)
mean=statistics.mean(savgol)
- print (mean)
- quit(-1)
start1 = np.datetime64(paiva1)
end1 = np.datetime64(paiva2)
start2 = np.datetime64(taika1)
end2 = np.datetime64(taika2)
- days1 = np.linspace(start1.astype('f8'), end1.astype('f8'), dtype='<M8[D]')
- napapaiva1 = np.datetime64("2020-04-01")
timedelta1= np.timedelta64(len(cases),'D')
- napapaiva2 = napapaiva1+timedelta1
- dada1 = np.linspace(napapaiva1.astype('f8'), napapaiva2.astype('f8'), dtype='<M8[D]')
days1 = pd.date_range(start1, end1, periods=len(cases)).to_pydatetime()
days2 = np.linspace(start2.astype('f8'), end2.astype('f8'), dtype='<M8[D]')
days3 = np.linspace(start1.astype('f8'), end2.astype('f8'), dtype='<M8[D]')
len1=len(days1)
len2=len(days2)
len3=len(days3)
x1 = np.linspace(0, len1, len1)
x2 = np.linspace(len1, len2, len2)
x3 = np.linspace(0, len3, len3)
lenx1=len(x1)
lenx2=len(x2)
lenx3=len(x3)
- print(days2)
- print (dates)
print (cases)
print (len(cases))
print (len2)
- quit(-1)
- print (len(dates))
- print (len(cases))
lencases1=len(cases)
- print(lencases1)
- quit(-1)
start1 = np.datetime64(paiva1)
days3 = np.linspace(start1.astype('f8'), end2.astype('f8'), dtype='<M8[D]')
meas=calcu_fourier_1(cases,days3)
logmeas=calcu_fourier_1(np.log(cases),days3)
expmeas=np.exp(logmeas)
alkumeas=savgol_filter(meas, 3, 2)
silitettymeas=meas
- days4=days3[:-1]
- nym=1
- for nym in range(0,10):
- start1=start1+np.timedelta64(nym,'D')
- days3 = np.linspace(start1.astype('f8'), end2.astype('f8'), dtype='<M8[D]')
- days4=days3[:-1]
#meas=calcu_fourier_1(cases,days3)
#plt.plot(days4,meas, color="red", alpha=0.3)
- plt.show()
- quit(-1)
- upper0=meas+devis
- lower0=meas-devis
- meas=silitettymeas
upper=meas+stdev
lower=meas-stdev
upper2=meas+stdev*2.4
lower2=meas-stdev*1.2
upper3=meas+stdev*0.5
lower3=meas-stdev*0.5
days4=days3[:-1]
- days4=days3
- plt.plot(days4,meas)
- plt.plot(days4,expmeas)
- plt.plot(days4,upper)
- plt.plot(days4,lower)
plt.fill_between(days4,upper2, lower2,color="#ffa0a0")
plt.fill_between(days4,upper, lower,color="#af8080")
plt.fill_between(days4,upper3, lower3,color="#8f6060")
plt.plot(days4,meas)
plt.scatter(days1,cases, color='k')
dateformat1 = mdates.DateFormatter('%d.%m')
plt.xticks(fontsize=16)
plt.yticks(fontsize=16, rotation=0)
ax1=plt.gca()
plt.title("Koronavirustapauksia - Fourier-ennuste", fontsize=18)
ax1.set_xlabel('Pvm', color='g',size=18)
ax1.set_ylabel('Koronatapauksia päivässä', color='#800000',size=18)
ax1.xaxis.set_major_formatter(dateformat1)
plt.ylim(0,ymax1)
plt.show()
print(".")
Licensing
[edit]- You are free:
- to share – to copy, distribute and transmit the work
- to remix – to adapt the work
- Under the following conditions:
- attribution – You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
- share alike – If you remix, transform, or build upon the material, you must distribute your contributions under the same or compatible license as the original.
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 08:25, 16 April 2022 |  | 892 × 520 (41 KB) | Merikanto (talk | contribs) | update |
| 08:42, 7 August 2021 | 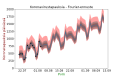 | 841 × 536 (43 KB) | Merikanto (talk | contribs) | update | |
| 08:54, 25 July 2021 |  | 906 × 557 (43 KB) | Merikanto (talk | contribs) | Update | |
| 07:46, 20 May 2021 |  | 977 × 507 (44 KB) | Merikanto (talk | contribs) | Update | |
| 07:36, 20 May 2021 | 956 × 367 (36 KB) | Merikanto (talk | contribs) | Upload | ||
| 08:25, 8 March 2021 |  | 799 × 528 (36 KB) | Merikanto (talk | contribs) | Update | |
| 18:09, 26 February 2021 |  | 576 × 432 (59 KB) | Merikanto (talk | contribs) | Upload | |
| 11:21, 25 January 2021 |  | 576 × 432 (46 KB) | Merikanto (talk | contribs) | Uploaded own work with UploadWizard |
You cannot overwrite this file.
File usage on Commons
There are no pages that use this file.
Metadata
This file contains additional information such as Exif metadata which may have been added by the digital camera, scanner, or software program used to create or digitize it. If the file has been modified from its original state, some details such as the timestamp may not fully reflect those of the original file. The timestamp is only as accurate as the clock in the camera, and it may be completely wrong.
| Width | 713.52pt |
|---|---|
| Height | 416.16pt |