Category:Proteins
Jump to navigation
Jump to search
biomolecule consisting of chains of amino acid residues | |||||
| Upload media | |||||
| Spoken text audio | |||||
|---|---|---|---|---|---|
| Instance of |
| ||||
| Subclass of |
| ||||
| Part of |
| ||||
| Has part(s) | |||||
| Different from | |||||
| |||||
Subcategories
This category has the following 193 subcategories, out of 193 total.
*
1
- 14-3-3 proteins (14 F)
?
- Unidentified proteins (3 F)
A
- Protein abundance (11 F)
- Actin-related protein 3 (10 F)
- Protein aggregation (2 F)
- Amino acid substitution (19 F)
- Amyloid (29 F)
- Amyloid beta-protein precursor (17 F)
- Anti-CRISPR proteins (6 F)
- Argonaute proteins (8 F)
- Armadillo domain proteins (20 F)
- Aurora B (4 F)
B
- Bcl-x protein (6 F)
- Beta amyloid production (10 F)
C
- Calgranulin A (2 F)
- Calgranulin B (2 F)
- Calgranulin C (1 F)
- Calmodulin-binding proteins (8 F)
- Protein classes (2 F)
- CLOCK proteins (9 F)
- Cofilin 1 (24 F)
- Protein conformation (236 F)
- Contractile proteins (171 F)
- Cortactin (13 F)
- Cyclin-dependent kinase 2 (6 F)
D
- Protein denaturation (13 F)
- Protein droplet (1 F)
E
- Encapsulins (2 F)
- Protein expression (10 F)
- Protein extraction (6 F)
F
- FAM63B (8 F)
- Fetal proteins (14 F)
G
- Glial fibrillary acidic protein (27 F)
- GM130 (11 F)
- Gtpase-activating proteins (1 F)
H
- Hemeproteins (12 F)
- Histatins (4 F)
- Homeodomain proteins (119 F)
- Protein homology detection (10 F)
- HSP20 heat-shock proteins (2 F)
I
- IGFBP1 (3 F)
- Integrin alpha2 (15 F)
- Protein isoforms (75 F)
K
L
- LIM-homeodomain proteins (8 F)
- Lithostathine (3 F)
M
- Mabinlin (3 F)
- Mad2 proteins (11 F)
- Matrix gla protein (6 F)
- Protein multimerization (166 F)
N
- Neoplasm proteins (41 F)
- Nerve tissue proteins (226 F)
- Neuregulin-1 (13 F)
- Nevit Dilmen Proteins (1 P, 215 F)
- Nodal protein (20 F)
- NPC1 (6 F)
- NPC1L1 (3 F)
O
- Obsolete protein structures (5 F)
- Opsonin proteins (13 F)
P
- Paxillin (27 F)
- Protein prenylation (12 F)
- Protein powders (19 F)
- Protein sequencing (5 F)
- Proteinuria (4 F)
- PrPC proteins (10 F)
- Pten phosphohydrolase (10 F)
Q
- Qa-SNARE proteins (9 F)
R
- R-SNARE proteins (6 F)
- Ran GTP-binding protein (6 F)
- Ras proteins (69 F)
- Repressor proteins (98 F)
- Resilins (7 F)
- RNA-protein interactions (19 F)
S
- Snare proteins (2 F)
- Protein stability (61 F)
- Protein subunits (51 F)
- SUMO protein (3 F)
- SUMO-1 protein (6 F)
- Protein synthesis (32 F)
T
- T-box domain proteins (16 F)
- Tacrolimus binding proteins (20 F)
- Telomere-binding proteins (18 F)
U
V
- Viral oncogene proteins (5 F)
W
Z
- Zyxin (6 F)
Media in category "Proteins"
The following 200 files are in this category, out of 1,375 total.
(previous page) (next page)-
-16 Frequency Polygon.JPG 710 × 610; 23 KB
-
005-CytochromecOxidase-cytox composite-1oco 1qle.tiff 1,660 × 810; 3.85 MB
-
022-PhotosystemI-reaction-centers.tiff 1,609 × 1,229; 5.67 MB
-
1-s.png 1,828 × 1,392; 2.23 MB
-
1-s2.0-S009286740900083X-gr1 lrg (2).jpg 1,477 × 1,362; 237 KB
-
10-nsp12 apo.png 1,542 × 1,528; 1.38 MB
-
11-nsp10.png 1,690 × 1,550; 1.11 MB
-
12-ndp10-sinefungin.png 1,730 × 1,440; 1.21 MB
-
12985 2016 561 Fig2 HTML.webp 778 × 579; 69 KB
-
13-nsp15.png 1,922 × 1,632; 1.31 MB
-
13197 2013 1042 Fig1 HTML.jpg 798 × 373; 42 KB
-
14-orf7a.png 1,296 × 1,450; 492 KB
-
15-orf7a.png 1,556 × 1,594; 1.14 MB
-
18 2014 1695 Fig6 HTML.webp 387 × 348; 17 KB
-
1a6d large.png 1,200 × 1,198; 581 KB
-
1a8y.jpg 1,000 × 797; 328 KB
-
1acb.png 737 × 789; 16 KB
-
1au1.jpg 1,000 × 797; 205 KB
-
1B34 SmD1 8strand.JPG 1,018 × 686; 87 KB
-
1b4r screenshot pkd DOMINIO.jpg 1,689 × 950; 113 KB
-
1bbp.jpg 1,000 × 797; 195 KB
-
1bbt.jpg 1,000 × 797; 265 KB
-
1brp.jpg 1,000 × 797; 202 KB
-
1bv1.png 526 × 491; 107 KB
-
1cd3.jpg 1,000 × 797; 302 KB
-
1csg.jpg 1,000 × 797; 103 KB
-
1dfn.jpg 1,000 × 797; 124 KB
-
1dxx.jpg 1,000 × 794; 311 KB
-
1e1w.jpg 1,000 × 794; 179 KB
-
1efn.png 1,280 × 1,280; 794 KB
-
1eot.jpg 1,000 × 797; 145 KB
-
1F6R.png 1,024 × 768; 282 KB
-
1f9j.jpg 1,000 × 797; 257 KB
-
1fga.jpg 1,000 × 794; 174 KB
-
1gcn.jpg 980 × 480; 35 KB
-
1gof front b.png 545 × 654; 68 KB
-
1gof front.GIF 545 × 654; 61 KB
-
1gog.jpg 864 × 792; 122 KB
-
1H NMR spectrum of Calmodulin.png 726 × 577; 26 KB
-
1i5j.jpg 1,000 × 797; 121 KB
-
1jxu.jpg 1,000 × 794; 146 KB
-
1K36.pdb.png 200 × 350; 28 KB
-
1k3i 7 bladed beta propeller of Galactose oxidase precursor.jpg 500 × 500; 47 KB
-
1L1O Replication protein A.png 784 × 600; 356 KB
-
1LG5-cilengitide.png 783 × 617; 152 KB
-
1lgn.jpg 1,000 × 797; 270 KB
-
1lj7.jpg 1,000 × 797; 306 KB
-
1n9c.jpg 1,220 × 768; 345 KB
-
1NEB.pdb.png 1,320 × 722; 194 KB
-
1omf.jpg 1,000 × 794; 342 KB
-
1pdg.jpg 1,000 × 797; 101 KB
-
1poh.jpg 1,000 × 797; 172 KB
-
1QNF Photolyase 190721.jpg 3,264 × 1,824; 1.31 MB
-
1ryo.jpg 1,000 × 749; 159 KB
-
1sbf.jpg 1,000 × 794; 298 KB
-
1snx.png 512 × 474; 45 KB
-
1stf.jpg 1,000 × 797; 139 KB
-
1svc NFkB DNA.png 800 × 800; 208 KB
-
1T5R Protein Leukocidin S.png 2,048 × 1,156; 332 KB
-
1T6C PPX.png 640 × 480; 153 KB
-
1T7P Crystal Structure.jpg 995 × 1,039; 263 KB
-
1tap.jpg 1,000 × 797; 134 KB
-
1tup.jpg 1,000 × 797; 279 KB
-
1UVQ.png 1,440 × 1,080; 464 KB
-
1v4l.jpg 1,000 × 794; 275 KB
-
1www.jpg 1,000 × 797; 251 KB
-
1xa9.jpg 1,000 × 794; 304 KB
-
1z2c asr r 500.jpeg 500 × 500; 46 KB
-
1Z3U.pdb1.jpg 550 × 550; 50 KB
-
1Z90 monomer.jpg 1,011 × 760; 61 KB
-
1Z90 oligomer.jpg 1,011 × 760; 417 KB
-
1ztm.jpg 1,000 × 797; 201 KB
-
2-S.png 2,250 × 1,572; 1.44 MB
-
2000 Antifreeze Proteins by Bill Nye for National Science Foundation.oga 1 min 3 s; 508 KB
-
2020 08 28 N glikan z legenda.png 3,000 × 2,100; 341 KB
-
2BC4.pdb1.png 1,436 × 759; 274 KB
-
2cb8.jpg 1,000 × 797; 156 KB
-
2cs4 PDB n-terminal domain chromosome 12 ORF 2.png 454 × 420; 123 KB
-
2fjz app.png 465 × 694; 70 KB
-
2gf1.jpg 1,000 × 797; 145 KB
-
2gtl.jpg 1,000 × 797; 323 KB
-
2gtl2.jpg 1,000 × 797; 312 KB
-
2gu0.jpg 1,000 × 794; 304 KB
-
2GVD.gross.jpg 581 × 631; 82 KB
-
2hmb.jpg 1,000 × 794; 180 KB
-
2HNX.png 960 × 720; 215 KB
-
2izi.jpg 1,000 × 797; 266 KB
-
2jsj obestatin.png 4,000 × 3,621; 1.12 MB
-
2k2o.png 410 × 527; 83 KB
-
2M9V asym r 500.jpg 500 × 500; 56 KB
-
2mhr.jpg 1,000 × 797; 219 KB
-
2p0a.jpg 1,000 × 794; 328 KB
-
2PTN.gif 1,014 × 720; 2.47 MB
-
2QH7.png 960 × 720; 240 KB
-
2ql1.jpg 1,000 × 794; 223 KB
-
2r5k.jpg 1,000 × 797; 294 KB
-
2rox.jpg 1,000 × 749; 170 KB
-
2roxa.jpg 1,000 × 749; 191 KB
-
2tmv.jpg 1,000 × 797; 376 KB
-
2vjt.jpg 1,000 × 794; 329 KB
-
2WINSurfaceRendering.png 640 × 515; 338 KB
-
2ygd.jpg 1,000 × 797; 332 KB
-
2yl2.png 314 × 349; 108 KB
-
2zd0.jpg 1,000 × 800; 267 KB
-
2zn9.jpg 1,000 × 794; 250 KB
-
3-nsp3.png 1,736 × 1,336; 1.01 MB
-
3c3v.jpg 1,000 × 797; 368 KB
-
3chy flavodoxin fold.png 1,000 × 875; 268 KB
-
3D protein.jpg 825 × 1,125; 231 KB
-
3D protein.png 400 × 400; 19 KB
-
3D structure of zingibain enzyme.png 1,101 × 656; 407 KB
-
3dkt.jpg 1,000 × 797; 402 KB
-
3FFN background removed.png 1,265 × 919; 483 KB
-
3jd6.jpg 1,000 × 797; 340 KB
-
3LFM FAT Mass and Obesity Associated (Fto) Protein.png 5,911 × 3,325; 2.94 MB
-
3llp.jpg 1,000 × 797; 271 KB
-
3mge.jpg 1,000 × 797; 275 KB
-
3s4eskrp1-es.jpg 676 × 514; 95 KB
-
3trx.jpg 1,000 × 794; 201 KB
-
3ux4.jpg 1,000 × 797; 267 KB
-
3wx6.jpg 1,000 × 797; 288 KB
-
4-6w9c.png 1,682 × 1,602; 1.11 MB
-
4dpv.jpg 1,000 × 797; 404 KB
-
4ldd.jpg 1,000 × 797; 186 KB
-
4qyz.jpg 1,000 × 797; 316 KB
-
4tsv bio r 250.jpg 250 × 250; 18 KB
-
4uft.jpg 1,000 × 797; 364 KB
-
5-nsp5.png 2,036 × 1,186; 1.36 MB
-
5cjf assembly 1.png 500 × 500; 100 KB
-
5cna.jpg 1,000 × 797; 274 KB
-
5exy.jpg 1,000 × 797; 354 KB
-
5flu.jpg 1,000 × 797; 300 KB
-
5jzc.jpg 1,000 × 797; 302 KB
-
5mke.jpg 1,000 × 797; 384 KB
-
5mug.jpg 1,000 × 797; 373 KB
-
5tio.jpg 1,000 × 797; 317 KB
-
5xqi.jpg 1,000 × 797; 330 KB
-
5zm8.jpg 1,000 × 797; 310 KB
-
6-nsp5.png 2,032 × 1,722; 1.57 MB
-
6ayf.jpg 1,000 × 797; 352 KB
-
6eho.jpg 1,000 × 797; 374 KB
-
6wbv.jpg 1,400 × 774; 323 KB
-
7-nsp7-8.png 1,798 × 1,080; 699 KB
-
7ACN.jpg 775 × 650; 463 KB
-
7ash.jpg 1,120 × 774; 464 KB
-
7b9s.jpg 900 × 774; 308 KB
-
7zva assembly-1.jpg 500 × 500; 60 KB
-
8-6w4b.png 2,042 × 1,562; 1,002 KB
-
8ohm.jpg 1,000 × 797; 236 KB
-
8tim TIM barrel topview.png 870 × 791; 253 KB
-
8tim TIM barrel.png 940 × 808; 328 KB
-
9-nsp12.png 1,868 × 1,622; 1.64 MB
-
9ilb.jpg 1,000 × 797; 204 KB
-
A protein interaction network in mESCs..jpg 600 × 362; 109 KB
-
A1BGgeneneighb.fcgi.png 485 × 75; 1,007 bytes
-
A1BGprod.fcgi.png 540 × 68; 1 KB
-
AA Rab3D.jpg 454 × 225; 20 KB
-
AAMDC.png 2,100 × 2,100; 1.8 MB
-
Aars alignment.png 5,262 × 2,771; 3.87 MB
-
ABCG2 with plasma membrane.jpg 586 × 668; 92 KB
-
Abundance over time.jpg 735 × 545; 57 KB
-
Abundància de integrina alfa 1 en els teixits humans.jpg 2,471 × 1,517; 716 KB
-
ActA Function.jpg 550 × 326; 60 KB
-
Acétyl-coenzyme A acétyltransférase.gif 945 × 637; 8.04 MB
-
ADAR tree.png 778 × 753; 149 KB
-
ADN et homéodomaine.png 571 × 644; 193 KB
-
AF-Q6UWT2-F1.png 426 × 213; 13 KB
-
Aflibercept (Eylea®).svg 480 × 350; 77 KB
-
Agrincartoon.png 640 × 480; 61 KB
-
Agrincartoon2.png 430 × 325; 59 KB
-
Ah2 nih.PNG 277 × 285; 53 KB
-
AICAR transformylase active site.png 776 × 623; 311 KB
-
AIMP2.png 2,800 × 2,800; 6.04 MB
-
AimR bound to a DNA Operator in SPbeta Phages.png 3,076 × 2,403; 1.13 MB
-
AK molmol1.jpeg 1,000 × 975; 104 KB
-
AKAP2.png 300 × 300; 31 KB
-
Alfa 2-antiplasmina.png 1,100 × 280; 55 KB
-
Alphafold 3D c2orf80 protein.png 742 × 584; 104 KB
-
Alphavbeta3 integrin expression in tumors.png 1,015 × 667; 591 KB
-
ALT structure.png 1,296 × 1,887; 659 KB
-
Amidinotransferase structure diagram.JPG 484 × 582; 66 KB
-
Amino Acid Multiplets in GOLGA8H.png 583 × 282; 55 KB
-
Amino acids by property.jpg 2,000 × 2,000; 1.19 MB
-
Aminoàcids 1-208 domini PZP AF-10 (versió b).gif 1,200 × 675; 13.1 MB
-
Aminoàcids 1-208 domini PZP AF-10.gif 1,200 × 675; 11.75 MB
-
Aminoàcids 699-782 AF-10.gif 1,200 × 675; 10.51 MB
-
Aminoàcids 704-779 AF-10 (versió b).gif 1,280 × 720; 9.34 MB
-
Aminoàcids 704-779 AF-10.gif 1,200 × 675; 7.06 MB
-
Anakinra.png 588 × 676; 18 KB
-
Analytical.jpg 640 × 400; 12 KB
-
AnalyticalChipExample.png 250 × 200; 7 KB
-
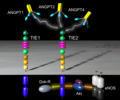
Ang-TIE-signaling.png 2,160 × 1,800; 969 KB
-
Anillin.jpg 1,124 × 454; 57 KB